This vignette demonstrates how to use
ddbs_interpolate_aw() to perform areal-weighted
interpolation. This technique is essential when you need to transfer
attributes from one set of polygons (source) to another incongruent set
of polygons (target) based on their area of overlap.
ddbs_interpolate_aw() can be 9-30x faster than
sf::st_interpolate_aw or areal::aw_interpolate
(see benchmarks in {ducksf} where prototype of
ddbs_interpolate_aw() was originally developed).
duckspatial handles these heavy geometric calculations
efficiently using DuckDB. We will cover three scenarios:
- Extensive vs. Intensive: Understanding the difference between mass-preserving counts and densities.
-
Fast Output: Returning a
tibble(no geometry) for maximum speed. - Database Mode: Performing operations on persistent database tables.
Setup Data
We will use the North Carolina dataset from the sf
package as our source, and create a generic grid as our
target.
Note: We project the data to Albers Equal Area (EPSG:5070) because accurate interpolation requires an equal-area projection.
library(duckspatial)
library(sf)
# 1. Load Source Data (NC Counties)
nc <- st_read(system.file("shape/nc.shp", package = "sf"), quiet = TRUE)
# 2. Transform to projected CRS (Albers) for accurate area calculations
nc <- st_transform(nc, 5070)
# 3. Create a Target Grid
grid <- st_make_grid(nc, n = c(10, 5)) |> st_as_sf()
# 4. Create Unique IDs (Required for interpolation)
nc$source_id <- 1:nrow(nc)
grid$target_id <- 1:nrow(grid)1) Extensive vs. Intensive Interpolation
Areal interpolation works differently depending on the nature of the data.
Case A: Extensive Variables (Counts)
Variables like population counts or total births (BIR74)
are spatially extensive. If a source polygon is split
in half, the count should also be split in half. We use
weight = "total" to ensure strict mass preservation
relative to the source.
# Interpolate Total Births (Extensive)
res_extensive <- ddbs_interpolate_aw(
target = grid,
source = nc,
tid = "target_id",
sid = "source_id",
extensive = "BIR74",
weight = "total",
output = "sf"
)
#> ✔ Query successfulVerification: The total sum of births in the result should match the original data (mass preservation).
Case B: Intensive Variables (Densities/Ratios)
Variables like population density or infection rates are
spatially intensive. If a source polygon is split, the
density remains the same in both pieces. duckspatial
handles this by calculating the area-weighted average.
# Interpolate 'BIR74' treating it as an intensive variable (e.g. density assumption)
res_intensive <- ddbs_interpolate_aw(
target = grid,
source = nc,
tid = "target_id",
sid = "source_id",
intensive = "BIR74", # Treated as density here
weight = "sum", # Standard behavior for intensive vars
output = "sf"
)
#> ✔ Query successfulVisual Comparison
Notice the difference in patterns. Extensive interpolation accumulates values based on how much “stuff” falls into a grid cell, while intensive interpolation smoothes the values based on overlap.
# Combine for plotting
plot_data <- res_extensive[, "BIR74"]
names(plot_data)[1] <- "Extensive_Count"
plot_data$Intensive_Value <- res_intensive$BIR74
plot(plot_data[c("Extensive_Count", "Intensive_Value")],
main = "Interpolation Methods Comparison",
border = "grey90",
key.pos = 4)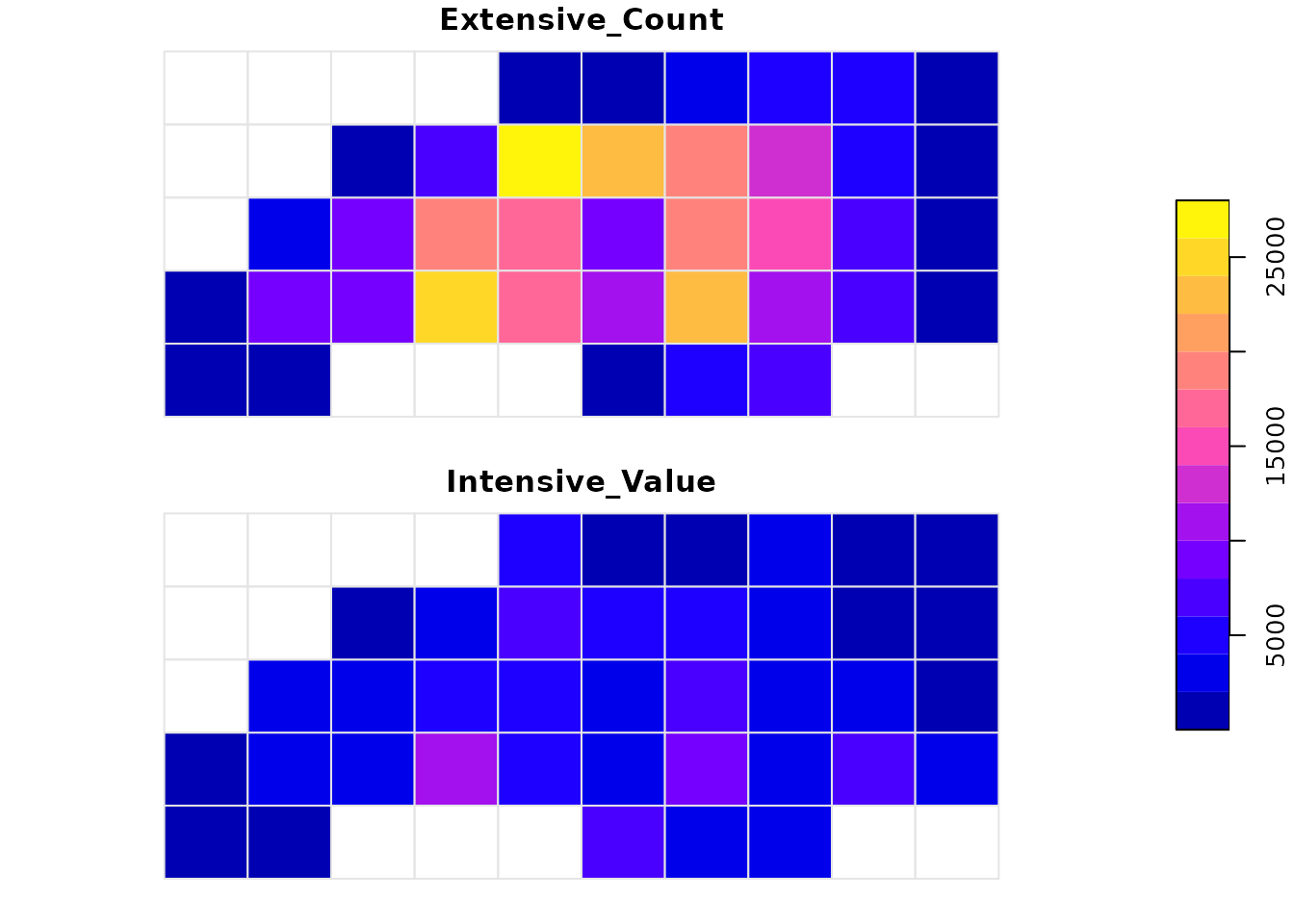
2) High Performance: Output as Tibble
If you are working with massive datasets, constructing the geometry
for the result sf object can be slow. If you only need the
interpolated numbers, set output = "tibble". This skips the
geometry construction step and is significantly faster.
# Return a standard data.frame/tibble without geometry
res_tbl <- ddbs_interpolate_aw(
target = grid,
source = nc,
tid = "target_id",
sid = "source_id",
extensive = "BIR74",
output = "tibble"
)
#> ✔ Query successful
head(res_tbl)
#> # A tibble: 6 × 3
#> target_id crs_duckspatial BIR74
#> <int> <chr> <dbl>
#> 1 1 EPSG:5070 1168.
#> 2 2 EPSG:5070 379.
#> 3 6 EPSG:5070 753.
#> 4 7 EPSG:5070 5731.
#> 5 8 EPSG:5070 8000.
#> 6 11 EPSG:5070 1417.3) Database Mode: Large Data Workflows
For datasets larger than memory, or for persistent pipelines, you can perform the interpolation directly inside the DuckDB database without pulling data into R until the end.
First, let’s establish a connection and load our spatial layers into tables.
# Create connection
conn <- ddbs_create_conn()
# Write layers to DuckDB
ddbs_write_vector(conn, nc, "nc_table", overwrite = TRUE)
#> ℹ Table <nc_table> dropped
#> ✔ Table nc_table successfully imported
ddbs_write_vector(conn, grid, "grid_table", overwrite = TRUE)
#> ℹ Table <grid_table> dropped
#> ✔ Table grid_table successfully importedNow we run the interpolation by referencing the table names. We can
also use the name argument to save the result directly to a
new table instead of returning it to R.
# Run interpolation and save to new table 'nc_grid_births'
ddbs_interpolate_aw(
conn = conn,
target = "grid_table",
source = "nc_table",
tid = "target_id",
sid = "source_id",
extensive = "BIR74",
weight = "total",
name = "nc_grid_births", # <--- Writes to DB
overwrite = TRUE
)
#> ℹ Table <nc_grid_births> dropped
#> ✔ Query successful
# Verify the table was created
DBI::dbListTables(conn)
#> [1] "grid_table" "nc_grid_births" "nc_table"We can now query this table or read it back later.
# Read the result back from the database
final_sf <- ddbs_read_vector(conn, "nc_grid_births")
#> ✔ table nc_grid_births successfully imported.
head(final_sf)
#> Simple feature collection with 6 features and 2 fields
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: 1054293 ymin: 1348021 xmax: 1677656 ymax: 1484503
#> Projected CRS: NAD83 / Conus Albers
#> target_id BIR74 x
#> 1 1 1168.3093 POLYGON ((1054293 1348021, ...
#> 2 2 378.5281 POLYGON ((1132214 1348021, ...
#> 3 6 752.9156 POLYGON ((1443895 1348021, ...
#> 4 7 5731.0103 POLYGON ((1521815 1348021, ...
#> 5 8 7999.6957 POLYGON ((1599735 1348021, ...
#> 6 11 1416.5579 POLYGON ((1054293 1416262, ...If we only wanted the database table, without the geometry, we could do:
ddbs_interpolate_aw(
conn = conn,
target = "grid_table",
source = "nc_table",
tid = "target_id",
sid = "source_id",
extensive = "BIR74",
weight = "total",
name = "nc_grid_births", # <--- Writes to DB
overwrite = TRUE,
output = "tibble"
)
#> ℹ Table <nc_grid_births> dropped
#> ✔ Query successfulAnd preview this table directly in the database:
DBI::dbGetQuery(conn, "SELECT * FROM nc_grid_births LIMIT 5")
#> target_id crs_duckspatial BIR74
#> 1 1 EPSG:5070 1168.3093
#> 2 2 EPSG:5070 378.5281
#> 3 6 EPSG:5070 752.9156
#> 4 7 EPSG:5070 5731.0103
#> 5 8 EPSG:5070 7999.6957